Get 1000 Hours free
On the UCSD Supercomputer
Start Your Trial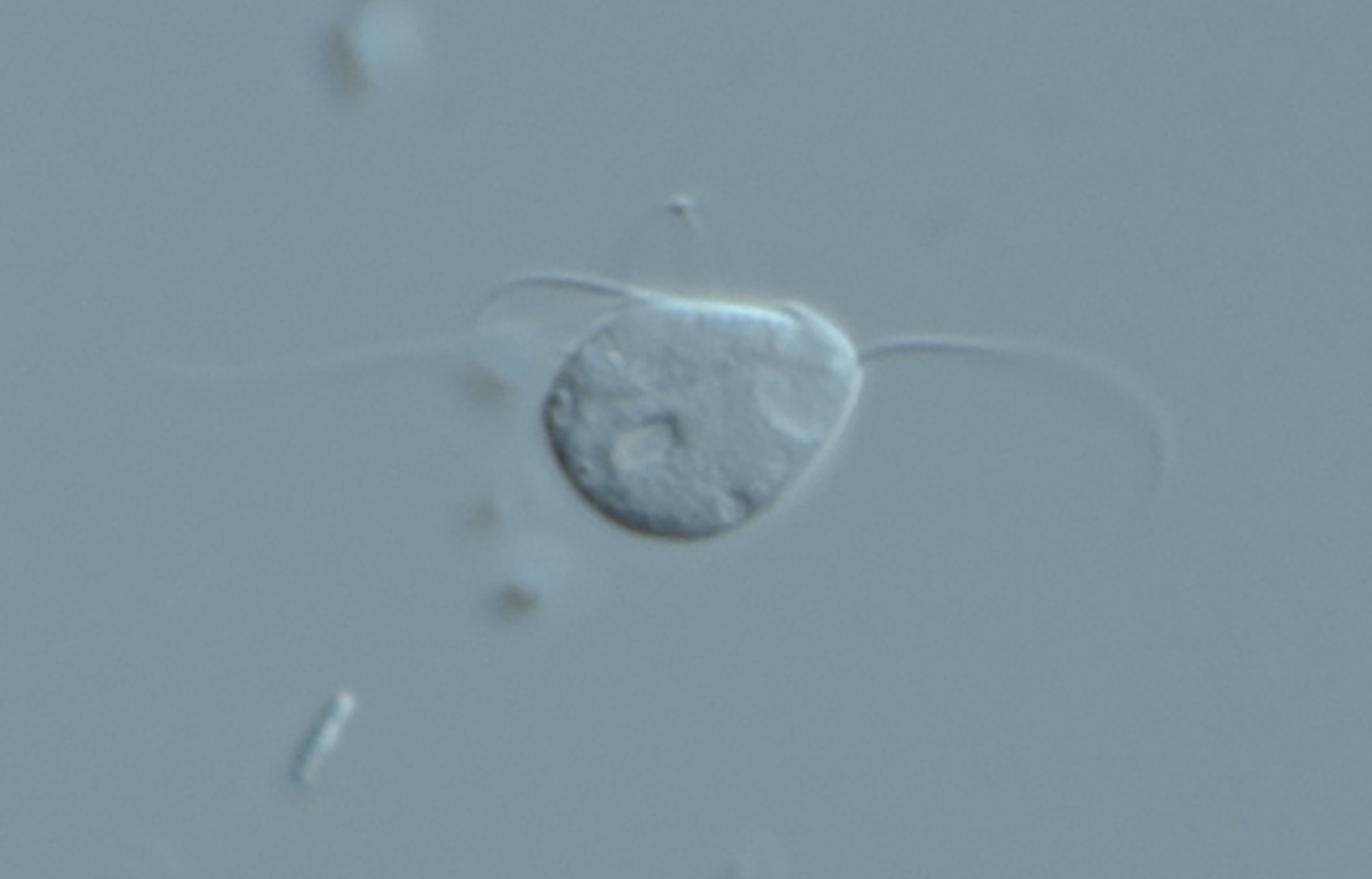
Genomics of Preaxostyla Flagellates Illuminates the Path Towards the Loss of Mitochondria
Novak and coworkers
The mitochondrion was long considered to be one of the defining elements of eukaryotic life. While it is well known that eukaryote species inhabiting low oxygen environments may have diminished mitochondrial complexity (and sometimes toss the mitochondrial respiratory function altogether), the vestigial machinery contained in Mitochondrion-Related Organelles (MRO) was thought to be required for eukaryotic life. Even though radically degenerate MROs (i.e. hydrogenosomes) have no role whatsoever in energy metabolism, the MRO provides other essential functions to eukaryotic cells.
Everything changed in 2016, when Karnkowska and colleagues reported a unicellular oxymonad (Monocercomonoides exilis; a chinchilla gut symbiont, Figure 1) that had lost all evidence of its mitochondrion. That is, all essential capabilities provided by the MRO were replaced through acquisition of alternative activities or became unnecessary. The authors noted that M. exilis had replaced one key mitochondrial feature, iron-sulfur cluster assembly, with a cytosolic sulfur mobilization system recruited from bacteria through lateral gene transfer. But many questions remain about the evolutionary events that allowed this organism to shuck its MRO completely.
Novak and coworkers (https://doi.org/10.1371/journal.pgen.1011050) recently shed new light on these events through extensive study of symbiotic and free-living Preaxostyla genomes. The authors did high quality genomic sequencing of a second oxymonad (Blattamonas nauphoetae) and a free-living anaerobe Paratrimastix pyriformis, which is sister to the oxymonads and compared the new DNA sequences with publicly available sequences from oxymonads M. exilis, and Streblomastix strix, and the free-living Trimastix marina. The origins of amitochondrial life were probed by extensive characterization of mitochondrion hallmark proteins, followed by comparisons of gene repertoires and metabolic functions. The authors relied on CIPRES to conduct their phylogenetic explorations using RAxML and MrBayes.
Key Findings:
Conclusions:
Loss of all traces of the mitochondria in oxymonads means that even the diminished function(s) of the Preaxostyla MRO are no longer essential. This study provides a strong basis to explore how those functions have been replaced. All oxymonads have apparently acquired activities from bacterial sources that replace MRO functions in iron cluster synthesis. All oxymonads have lost the glycine cleavage system and serine hydroxymethyl transferase system, but it remains unclear how the oxymonads compensate for the loss. Have oxymonads lost the ability to create S-adenosyl-methionine, or do they compensate for loss of the GCS system through a cytosolic alternative? There is provocative evidence they may have recruited a eukaryotic GCS-L2 protein that could play a role. Further study will be required to fully understand the loss of peroxisomal marker Pex3 and its potential role in the free-living Paratrimastix, and the study provides a significant opportunity to explore Golgi infrastructure and function in the Preaxostyla.
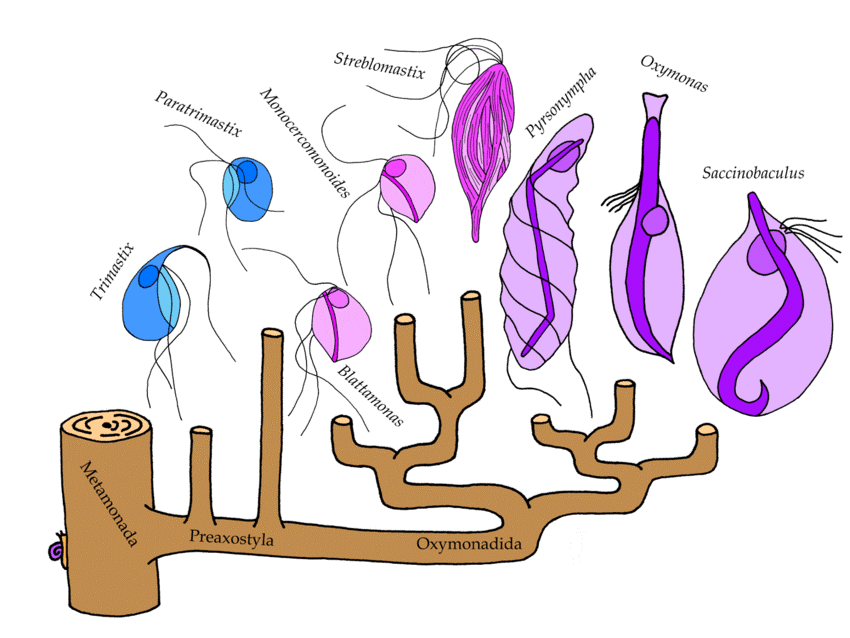
Novak and coworkers recently shed new light on these events through extensive study of symbiotic and free-living Preaxostyla genomes. The authors did high quality genomic sequencing of a second oxymonad (Blattamonas nauphoetae) and a free-living anaerobe Paratrimastix pyriformis, which is sister to the oxymonads and compared the new DNA sequences with publicly available sequences from oxymonads M. exilis, and Streblomastix strix, and the free-living Trimastix marina. The origins of amitochondrial life were probed by extensive characterization of mitochondrion hallmark proteins, followed by comparisons of gene repertoires and metabolic functions. The authors relied on CIPRES to conduct their phylogenetic explorations using RAxML and MrBayes. The resulting phylogenetic relationships among the Preaxostyla flagellates are shown in the image above, where free living members are shown in blue and the symbiotic Oxymonads are shown in purple.
Key Findings
Conclusions:
Loss of all traces of the mitochondria in oxymonads means that even the diminished function(s) of the Preaxostyla MRO are no longer essential. This study provides a strong basis to explore how those functions have been replaced. All oxymonads have apparently acquired activities from bacterial sources that replace MRO functions in iron cluster synthesis. All oxymonads have lost the glycine cleavage system and serine hydroxymethyl transferase system, but it remains unclear how the oxymonads compensate for the loss. Have oxymonads lost the ability to create S-adenosyl-methionine, or do they compensate for loss of the GCS system through a cytosolic alternative? There is provocative evidence they may have recruited a eukaryotic GCS-L2 protein that could play a role. Further study will be required to fully understand the loss of peroxisomal marker Pex3 and its potential role in the free-living Paratrimastix, and the study provides a significant opportunity to explore Golgi infrastructure and function in the Preaxostyla.
